Getting the (Semantic) Sense out of a User Query
Contents
The BIOfid Semantic Search
Within the BIOfid-project, we create a semantic search portal (hereafter “BIOfid portal”) to help our users to access legacy biodiversity literature more easily. Hence, since the BIOfid portal has a deeper “understanding” of both the texts and the included species, it allows the users to get more relevant documents. Moreover, the BIOfid portal interprets the user query and transform it ad hoc into a graph database query, to learn more about their intention.
To do so, the BIOfid portal processes every query as follows:
- Query Preprocessing (Cleaning, Simple Semantics Extraction)
- Natural Language Processing (NLP) applying Machine Learning
- NLP interpretation and conversion to a SPARQL query and running it against a graph database
- Converting the graph database response to a document database query and running the query
- Prepare the retrieved documents for user presentation
In this post, I will go into detail on the steps 1 to 3.
Please note that although this text is written in English, at the time of writing, the BIOfid portal only interprets user queries in german language!
Query Preprocessing
After the user query is handed to the BIOfid portal, there are some cleaning steps first. For example, I had the impression that the NLP result is worse, when a sentence does not end with a question mark ("?") or a full stop ("."). Since a search engine query usually does not contain such a thing, a full stop will be added to all queries missing either.
Also, if there are very characteristic keywords included in the query they are excluded from the query before the NLP. These characteristic keywords are e.g. ‘since’ followed by four digits hinting for a restriction of the time span. Regular Expressions are pretty useful for simple string manipulation/extraction and take away a lot of work from the post-processing of the NLP.
Furthermore, since this is a biological search, I had to consider queries for species. The problem is that a biologist can write the same species in different ways. For example, the following user queries (for the Common beech) should result in the (roughly) same result set:
- Fagus sylvatica
- Fagus sylvatica L.
- Fagus sylvatica L., 1758
Although the NLP has specifically a model that should recognise multi-token1 species names, this is not reliable. Hence, the BIOfid portal will shrink down complex multi-token species names to just a single token. For example, “Fagus sylvatica L.” will be reduced to “Fagus” only. After the NLP ran, the “hidden” tokens are reinserted into the query for further processing. This approach allows the NLP to recognize that there is a species involved, but takes away the complexity to also handle all the additional words that do not add any information to the NLP.
Get the Semantics
Applying Natural Language Processing
The state of the art in NLP is using machine learning. Hence, the BIOfid portal connects to an NLP server2 that is doing all the heavy lifting and returns the (in our case) user query semantics. If you wonder how this output would look like, I suggest having a look at Displacy, which gives a very human-friendly representation of how an NLP result looks like (Fig. 1).
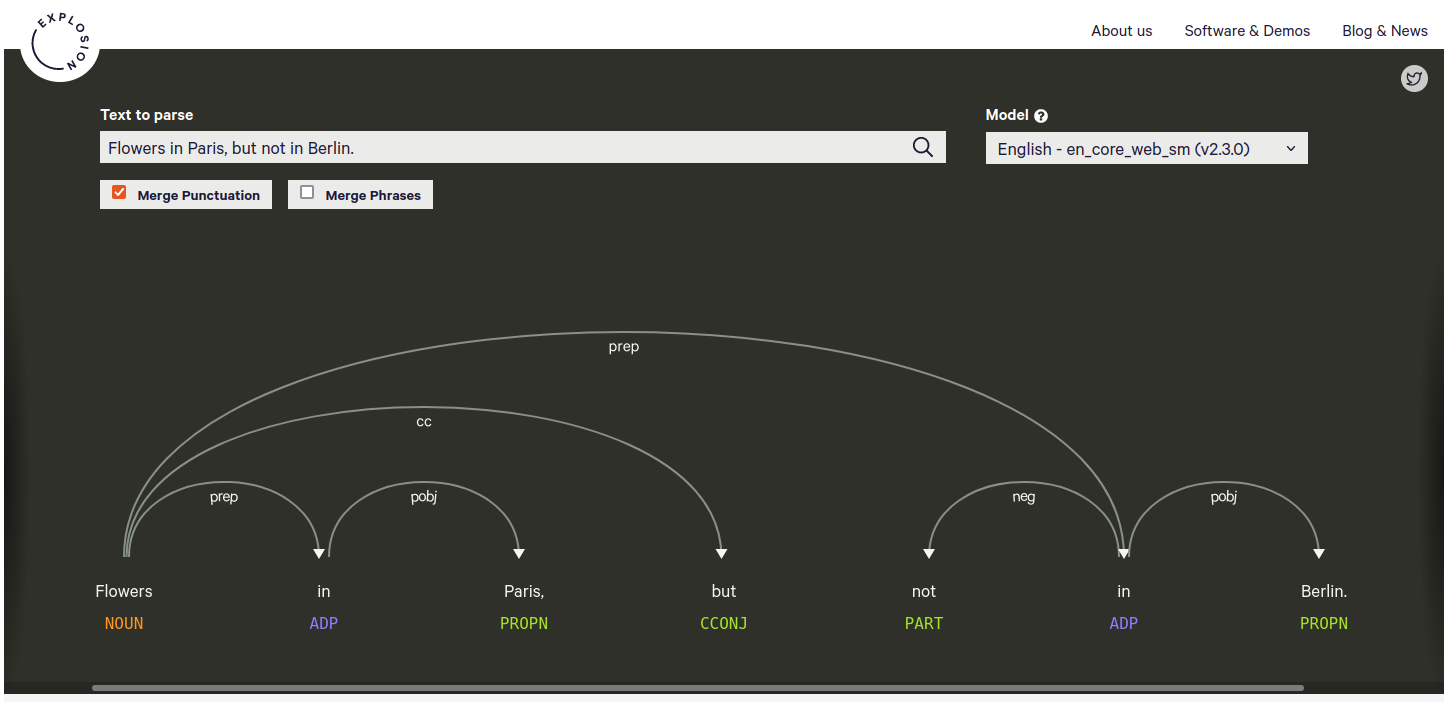
Figure 1
A human-friendly Natural Language Processing result for the query ‘Flowers in Paris, but not in Berlin’. Generated by Displacy. The labels below the words annotate the word’s part of speech. The arrows indicate word dependencies with the respective abbreviated dependency type.
The NLP returns basic information on the part of speech (i.e. is the word a noun, an adjective, a conjunction or something else?), the lemma (i.e. the basic form of the word, which would be “run” for the word “ran”), and (very important) the word dependency. The word dependency allows the BIOfid portal to make more sense from the user query and it drives the generation of the graph database query.
Extracting Word Dependencies
It is the word dependency, that is processed in the next step. The BIOfid portal runs through a large sequence of rules to extract triple statements from the word dependency information. For example, for the user query “Flowers in Paris, but not in Berlin” (see also Fig. 1), this would generate statements like this:
|
|
with PROPN meaning “Proper Noun”3.
At this point, the BIOfid portal has no idea what a flower is. But it knows that “flower” is an important word related to the locations “Paris” and “Berlin”. Wait!? Locations!? Yes! Thanks to another process in the NLP, the Named Entity Recognition, we now know that “Paris” and “Berlin” are locations and not e.g. plants4. Also, we know that the second statements is negated!
To tackle this task programmatically, my current implementation (for I knew not what I did) is a long if-else statement, which is a mess to maintain. The lesson learned from my current approach: I currently would recommend using either a pipeline in conjunction with a chain of responsibility or applying a rules design. The latter would probably make the intention more clear. Anyway, this task has to be approached in a maintainable manner, because you want to be able to expand the analytical properties easily!
Making the Semantics Unique
Although the BIOfid portal knows at this stage that a “flower” (whatever this may be) is related to two locations, it still does not know which locations exactly. “Flower”, “Paris”, and “Berlin” are still only strings! We have to provide the BIOfid portal a way to talk to the graph database and referring to these words with unique terms. For this, we apply Uniform Resource Identifiers (URIs). URIs are (most of the time) website addresses, because a web addresses are always unique.
Hence, the BIOfid portal will look up in an index database the URIs for each of the terms, providing additionally the semantic context. Both “Paris” and “Berlin” are locations. So, the index database uses both, the string (more specific: the lemma) of the name and the context “Location” to return URIs that fit these criteria.
Yes, this is not perfect and will probably still return URIs that are not correct in the context (probably you meant the Paris in France, not the one in Idaho, Maine, Denmark, or Canada). However, from a user query, it is very hard (if not impossible) to extract more meaningful information, without just ignoring certain results. In our case, the optimal result for the URI request per term would be like https://sws.geonames.org/2988507/ (Paris), https://www.geonames.org/2950159/ (Berlin), and http://purl.obolibrary.org/obo/PO_0009046 (flower).
Besides the URI, the index database also returns the position of this URI that it will take in a SPARQL triple. Should the URI be used as a predicate (position = 2) or as an object (position = 3). This allows for more sophisticated queries that include e.g. querying for flower colours. Such a query needs a term like https://www.biofid.de/bio-ontologies#has_petal_color, which is a predicate. Like in a query for “red flowers”5:
|
|
Generating the Graph Database Query
We are getting to the hard part! At this point, the BIOfid portal knows how relevant terms in the query interact and what their URI is. Both is needed for a SPARQL query, the query language many graph databases understand.
To make this transformation, I applied the jinja2 template engine. Hence, the BIOfid portal provides the template engine with the generated objects and their relations and the engine returns a complete SPARQL string. Of course, the engine needs templates to call. These templates will be a lot of if-else, because the single statements or the SPARQL brackets to insert/ignore are often very context sensitive.
To give you an idea of a template, here is a fraction of the template that get the systematics of a taxon:
|
|
You see that the template takes into account, if there is negation involved and if there are AND-related taxons in the query. To hide away some complexity, the include calls other templates and their content is injected at the respective position.
The whole thing is a beast! To develop and maintain such a complex code generator, I apply test-driven development! Before writing or editing any of the templates, I write a test. That is a user query input and the expected SPARQL string. If you want to be completely sure about your generated SPARQL query, you can also set up a small ontology, load it in an in-memory graph database(e.g. RDFLib in Python) and run the generated query against it. The templates become complex very fast and it is not possible to tame them otherwise.
That’s it! The BIOfid portal then throws the generated SPARQL to the graph database, which then returns its output. The BIOfid portal takes the graph database response and generates from it a document database request, while still acknowledging the relationships between the terms in the original user query.
But that is subject for another story!
-
A token is (most of the time) equivalent to a word (e.g. “beech”, “city”). Hence, a “multi-token” word would be something like “New York” or “Palma de Mallorca”. In our case, “Fagus sylvatica L.” is a three token word. ↩︎
-
Wahed Hemati, Tolga Uslu, Alexander Mehler: TextImager: a Distributed UIMA-based System for NLP. COLING (Demos) 2016: 59-63 [PDF] ↩︎
-
A proper noun (which is often also referred to as Named Entity), “is a real-world object, such as a person, location, organization, product, etc., that can be denoted with a proper name” (from Wikipedia; CC-BY-SA 3.0). A proper noun always is referring a single existing object. Hence, “Barack Obama” is a proper noun, because there is only one. “Leaf” is not a proper noun, because there are literally billions of them. ↩︎
-
This is actually quite tricky, because there is a genus of plants called Paris. But “Paris” is in general a word you do not want to screw with: https://en.wikipedia.org/wiki/Paris_(disambiguation) ↩︎
-
The URI for red is http://purl.obolibrary.org/obo/PATO_0000320. ↩︎
Last Modified on 2021-09-15.
